Epigenetic sequencing
Epigenetics is emerging as a key tool for understanding the mechanistic underpinnings that regulate gene expression. Monitoring methylation, DNA accessibility, DNA-protein interactions, and non-coding small RNA functions can help to further understand the molecular landscape of cellular function, particularly when coupled with RNA-seq and DNA-seq.
Methods of interrogating these epigenetic factors are described in the technical notes and other resources below.
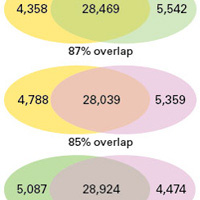
ChIP-seq libraries for transcription factors
Learn how researchers used our low-input ThruPLEX DNA-Seq kit to perform ChIP-seq with 10-fold less starting sample.
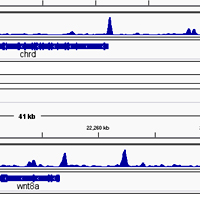
ChIP-seq libraries from single-stranded DNA
The ChIP Elute Kit and DNA SMART ChIP-seq kit are robust, reliable tools for low-input ChIP-seq applications.
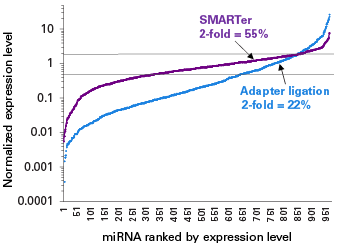
Full-length small RNA libraries
The SMARTer smRNA-Seq Kit for Illumina allows for analysis of diverse small RNA species with high accuracy and sensitivity.
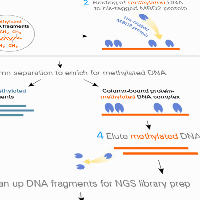
Methylated DNA-seq with MBD2
The EpiXplore Meth-Seq DNA Enrichment Kit provides a user-friendly solution for high-throughput analysis of DNA methylation patterns.

Profiling transcription factors with CUT&RUN sequencing
Learn how Dr. Karl Glastad of the University of Pennsylvania overcame the limits of ChIP-seq to identify the molecular determinants underpinning caste-differential gene expression in ants.
Learn moreTakara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.